Can Science Crack the KRAS Code?
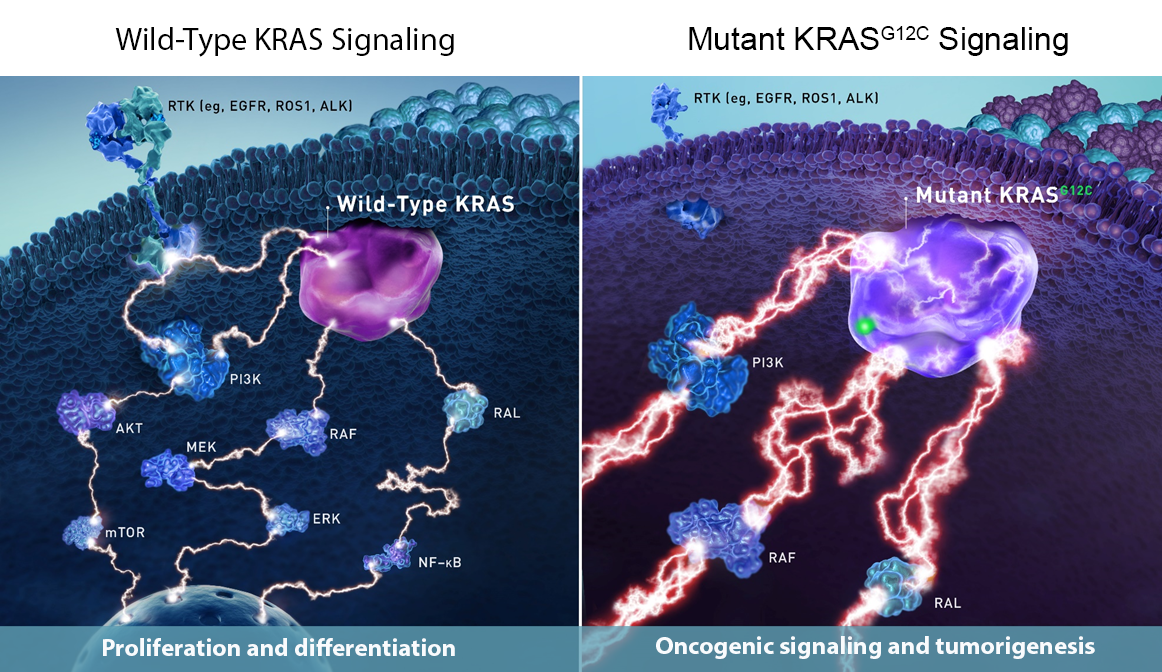
All human cells contain a gene called KRAS that serves as a key regulator of signaling pathways responsible for cell proliferation, differentiation, and survival. If a mutation occurs in a KRAS gene, it may allow cells to multiply out of control, which can cause cancer.
Scientists have known for decades that KRAS mutations are among the most common drivers of cancer because KRAS is the most frequently mutated oncogene, yet researchers have struggled to find a molecular “key” that can stop this out-of-control cell growth. A history of unsuccessful efforts to target KRAS led many to believe that KRAS mutations could be “undruggable.”
KRAS G12C is an oncogenic driver mutation in multiple cancer types
KRAS mutations play a role in some of the most common and deadly carcinomas, including lung, colorectal, and pancreatic cancers. One single type of KRAS mutation — called KRAS G12C — accounts for about 44% of all KRAS mutations. G12C is a single point mutation with a glycine-to-cysteine substitution at codon 12. This substitution favors the activated state of KRAS, amplifying signaling pathways that lead to oncogenesis.
KRAS G12C is particularly prevalent in non-small cell lung cancer (NSCLC), which makes up about 85% of all lung cancer cases in the U.S. Approximately 13% of Americans with NSCLC have the KRAS G12C mutation, and there are about 23,000 new cases of KRAS G12C NSCLC diagnosed every year in the U.S. alone. KRAS G12C is also found in 1-3% of colorectal and pancreatic cancer patients.
Despite high unmet need among patients with certain cancers, attempts by biotech researchers to inhibit KRAS directly have proven challenging. Many scientists have struggled to find surface targets on the protein where drugs could get a foothold to disrupt unchecked cell growth. And the inhibitors that some uncovered were non-selective, binding to both mutated and wild-type (non-mutated) KRAS, which can block the normal function of wild-type KRAS.
Targeting a unique surface groove on KRAS G12C
Recent scientific advancements revealed that the mutant cysteine KRAS G12C creates a narrow pocket that is susceptible to targeting. Researchers hypothesize that an adjacent histidine 95 (H95) residue may provide a site to stabilize drug-protein interactions. Using this binding pocket and H95, covalent inhibitors may lock KRAS G12C in the inactive state, blocking oncogenic signaling without disrupting the normal function of unmutated KRAS. This highly specific, targeted approach may also allow investigational use in combination with targeted and immunotherapies.
Advances in understanding the protein structure of KRAS have prompted further investigations and may provide insights into the role of KRAS mutations in cancer development and maintenance. Amgen is committed to investigating KRAS G12C for patients.
Amgen is currently enrolling for phase 1 and phase 2 clinical trials investigating KRAS G12C inhibitors.
To learn more, visit amgenoncology.com
REFERENCES
- National Cancer Institute. https://www.cancer.gov/types/lung/hp/non-small-cell-lung-treatment-pdq#_4. Accessed August 15, 2019.
- Bray F, et al. CA Cancer Res. 2018;68:394-424.
- American Cancer Society. https://www.cancer.org/cancer/lung-cancer.html. Accessed August 27, 2019.
- National Cancer Institute. https://seer.cancer.gov/statfacts/html/lungb.html. Accessed August 29, 2019.
- National Cancer Institute. https://seer.cancer.gov/archive/csr/1975_2015/results_merged/topic_survival.pdf. Accessed August 7, 2019.
- Skoulidis F, et al. Nat Rev Cancer. 2019;19:495-509.
- Chen Z, et al. Nat Rev Cancer. 2014;14:535-546.
- Ryan MB, et al. Nat Rev Clin Oncol. 2018;15:709-720.
- Biernacka A, et al. Cancer Genet. 2016;209:195-198.
- Stephen AG, et al. Cancer Cell. 2014;25:272-281.
- Cox AD, et al. Nat Rev Drug Discov. 2014;13:828-851.
- Shin Y, et al. ACS Chem Lett. 2019;11:1302-1308.